GenFam: gene family‐based classification and functional enrichment analysis
The online version available here
What is GenFam?
- GenFam is a new computational approach which allows annotation, classification, and enrichment of genes based on their gene family in plants genome. For example, GenFam can be useful to identify overrepresented differentially expressed genes obtained from RNA‐seq experiment (read GenFam paper for more details)
- GenFam and its integrated database comprises of 384 unique gene families and supports gene family analyses for 60 plant genomes.
- GenFam empowers users to readily interpret information of gene families (e.g., WRKY and bZIP) rather than GO terms or hierarchy alone in their queries, and move forward to selecting favorite overrepresented genes (or families) for downstream studies and interpretation.
- Here, I have described how to use GenFam as command line interface (CLI). The online version of GenFam can be found here
How to use GenFam as command line interface (CLI)?
- GenFam CLI is integrated into bioinfokit (v1.0.0 or later)
- Check How to install bioinfokit and usgae .
- Download test dataset for G.raimondii
GenFam classification and enrichment analysis for cotton genes
# I am using interactive python interpreter (Python 3.8.5)
# you must have active internet connection for GenFam analysis
>>> from bioinfokit.analys import genfam
# perform genfam analysis on downloaded G.raimondii dataset
>>> res = genfam()
>>> res.fam_enrich(id_file='grai_id.txt', species='grai', id_type=1)
# by default fam_enrich will use Fisher exact test and Benjamini-Hochberg FDR method
# get enriched gene families
>>> res.df_enrich
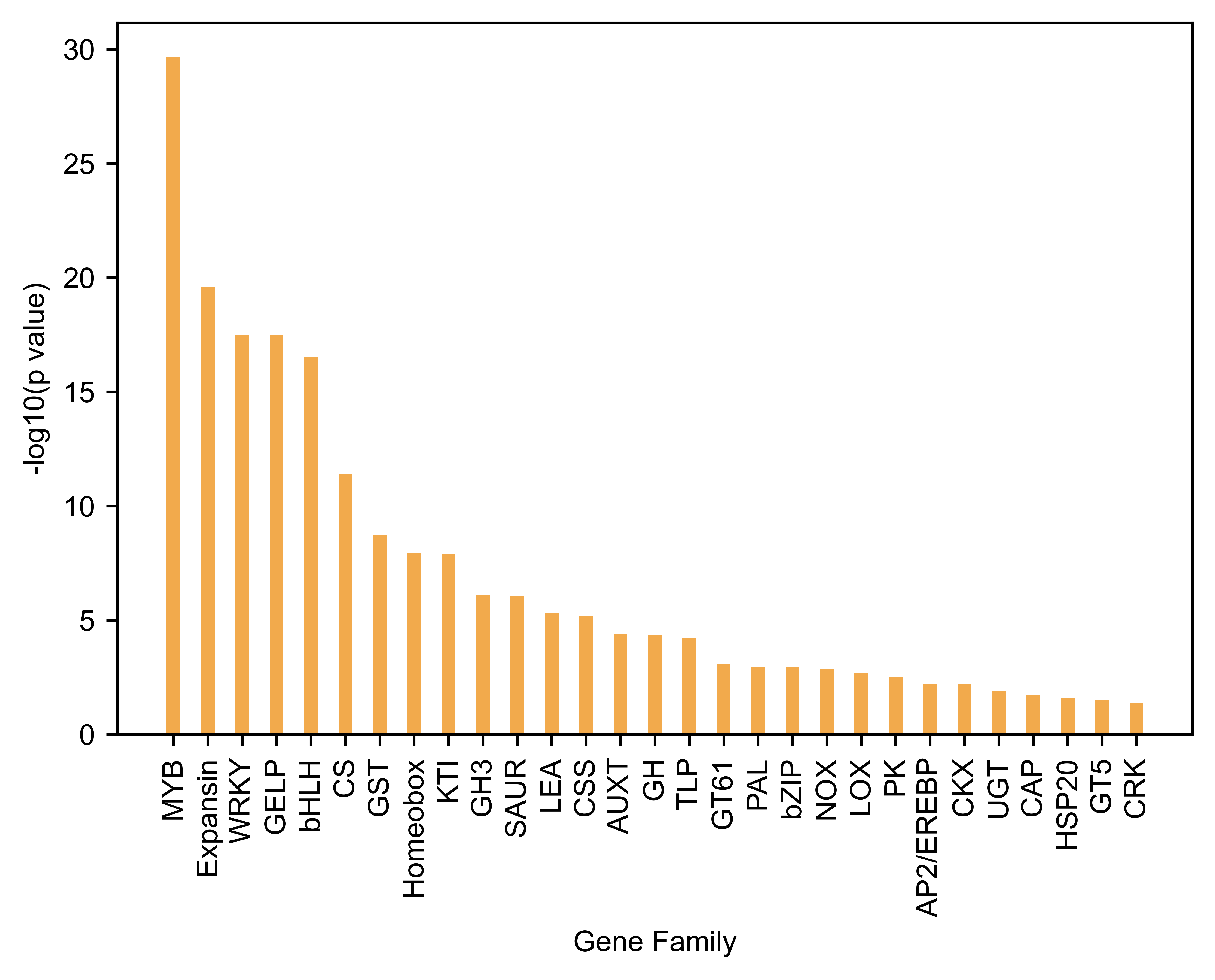
Gene Family Short name p value FDR
0 MYB gene family MYB 2.152548e-30 1.291529e-28
17 Expansin gene family Expansin 2.528412e-20 7.585235e-19
1 WRKY gene family WRKY 3.259473e-18 4.942421e-17
16 GDSL-like lipase (GELP) gene family GELP 3.294948e-18 4.942421e-17
2 Basic helix-loop-helix (bHLH) gene family gene... bHLH 2.889523e-17 3.467428e-16
13 Cellulose synthase (CS) gene family CS 4.071289e-12 4.071289e-11
14 Glutathione S-transferase (GST) gene family GST 1.786892e-09 1.531622e-08
4 Homeobox gene family Homeobox 1.131515e-08 8.310631e-08
20 Kunitz Trypsin Inhibitor (KTI) gene family KTI 1.246595e-08 8.310631e-08
24 GH3 gene family GH3 7.769853e-07 4.661912e-06
18 SAUR gene family SAUR 8.787804e-07 4.793347e-06
8 Late embryogenesis abundant (LEA) gene family LEA 4.981083e-06 2.490542e-05
12 Chalcone and stilbene synthase (CSS) gene family CSS 6.766939e-06 3.123203e-05
23 Auxin transport (AUXT) gene family AUXT 4.088460e-05 1.728157e-04
5 Glycoside hydrolase (GH) gene family GH 4.320394e-05 1.728157e-04
21 Thaumatin-like (TLP) gene family TLP 5.855903e-05 2.195964e-04
27 Glycosyltransferase 61 (GT61) gene family GT61 8.441974e-04 2.979520e-03
25 Phenylalanine ammonia lyase (PAL) gene family PAL 1.098768e-03 3.662560e-03
10 Basic leucine zipper (bZIP) gene family bZIP 1.192707e-03 3.766444e-03
11 NADPH oxidase (NOX) gene family NOX 1.357703e-03 4.073110e-03
19 Lipoxygenase (LOX) gene family LOX 2.056488e-03 5.875680e-03
28 Protein kinase (PK) gene superfamily PK 3.250643e-03 8.865390e-03
3 AP2/EREBP gene family AP2/EREBP 6.056923e-03 1.575365e-02
26 Cytokinin oxidase (CKX) gene family CKX 6.301461e-03 1.575365e-02
7 UDP-glycosyltransferases (UGT) gene family UGT 1.252574e-02 3.006178e-02
6 Cysteine-rich secretory proteins, Antigen 5, a... CAP 1.992329e-02 4.597683e-02
15 HSP20 gene family HSP20 2.606746e-02 5.792769e-02
9 Glycosyltransferase 5 (GT5) gene family GT5 3.031398e-02 6.495853e-02
22 Cysteine-rich receptor-like kinases (CRK) gene... CRK 4.199360e-02 8.688330e-02
# enriched gene families and all gene families along with GO annotation will be saved in
# fam_enrich_out.txt and fam_all_out.txt in the current working directory
# figure of enriched gene families (genfam_enrich.png) will also be saved
# get genfam analysis information
>>> res.genfam_info
Parameter Value
0 Total query gene IDs 652
1 Number of genes annotated 518
2 Number of enriched gene families (p < 0.05) 29
3 Plant species Gossypium raimondii v2.1
4 Statistical test for enrichment Fisher exact test
5 Multiple testing correction method Benjamini-Hochberg
6 Significance level 0.05
# if you are not sure about the type of gene IDs acceptable for GenFam, you can check it using check_allowed_ids
# function
>>> res.check_allowed_ids(species='grai')
Allowed ID types for Gossypium raimondii v2.1
Phytozome locus: Gorai.006G113100
Phytozome transcript: Gorai.013G113000.1
Phytozome pacID: 26782432
Enriched gene families figure,
GenFam classification and enrichment analysis for rice genes
- Download test dataset for O.sativa
>>> res.fam_enrich(id_file='osat_id.txt', species='osat', id_type=2)
>>> res.df_enrich
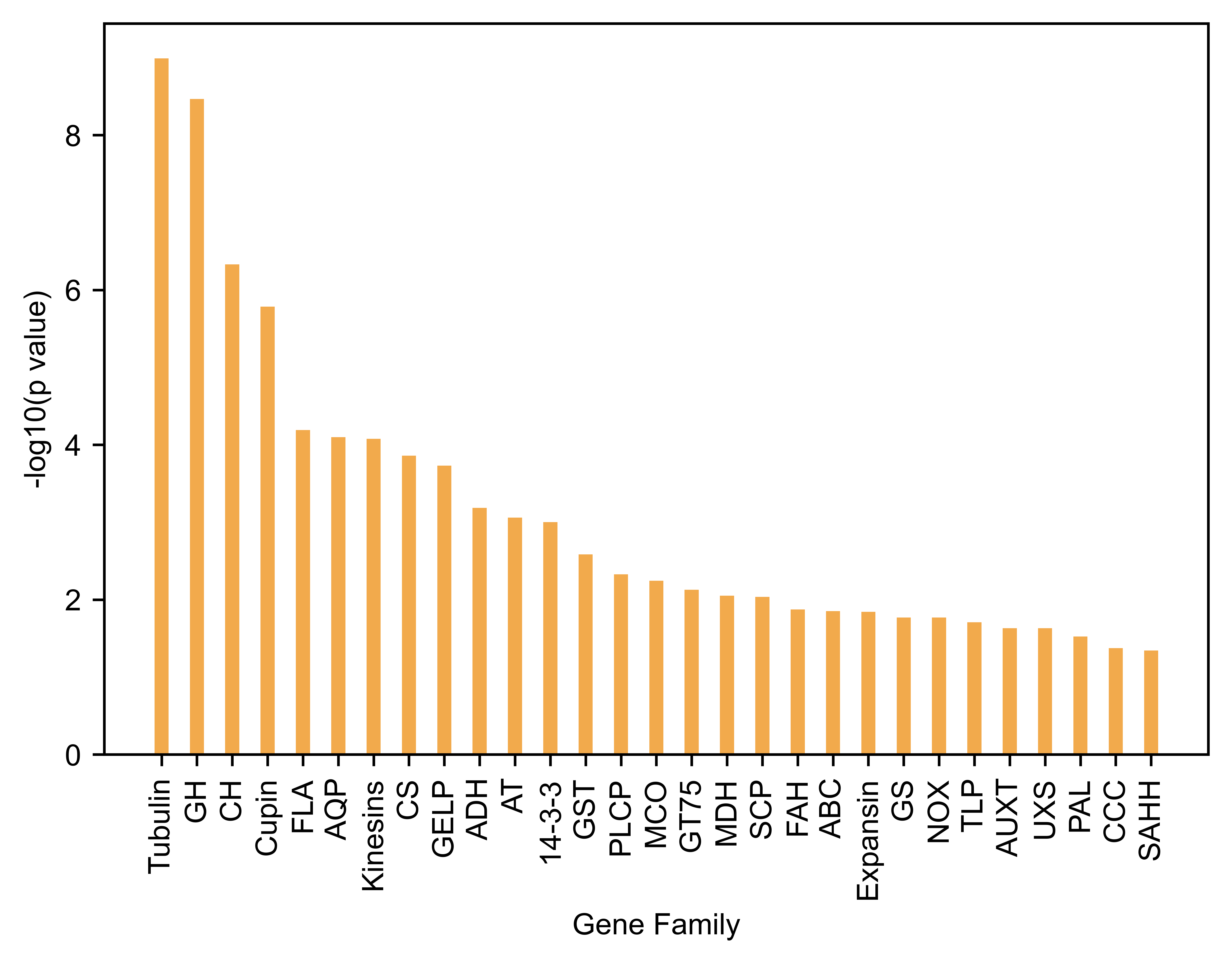
Gene Family Short name p value FDR
27 Tubulin gene family Tubulin 1.018099e-09 1.547510e-07
16 Glycoside hydrolase (GH) gene family GH 3.405050e-09 2.587838e-07
7 Core histone (CH) gene family CH 4.670238e-07 2.366254e-05
9 Cupin gene family Cupin 1.637162e-06 6.221217e-05
11 Fasciclin-like arabinogalactan (FLA) gene family FLA 6.453438e-05 1.814660e-03
4 Aquaporins (AQP) gene family AQP 7.987808e-05 1.814660e-03
18 Kinesins gene family Kinesins 8.356985e-05 1.814660e-03
6 Cellulose synthase (CS) gene family CS 1.377311e-04 2.616891e-03
13 GDSL-like lipase (GELP) gene family GELP 1.858720e-04 3.139171e-03
2 Aldehyde dehydrogenase (ADH) gene family ADH 6.513123e-04 9.899947e-03
3 Aminotransferase (AT) Gene Family AT 8.690746e-04 1.200903e-02
0 14-3-3 gene family 14-3-3 9.966784e-04 1.262459e-02
15 Glutathione S-transferase (GST) gene family GST 2.597434e-03 3.037000e-02
23 Papain-like cysteine proteases (PLCP) gene family PLCP 4.697927e-03 5.100606e-02
21 Multicopper oxidase (MCO) gene family MCO 5.686257e-03 5.762074e-02
17 Glycosyltransferase 75 (GT75) gene family GT75 7.438912e-03 7.066967e-02
19 Malate dehydrogenases (MDH) gene family MDH 8.891813e-03 7.773310e-02
25 Serine carboxypeptidase (SCP) gene family SCP 9.205236e-03 7.773310e-02
12 Fatty acid hydroxylase (FAH) gene family FAH 1.343143e-02 1.036782e-01
1 ABC transporter gene family ABC 1.402746e-02 1.036782e-01
10 Expansin gene family Expansin 1.432396e-02 1.036782e-01
14 Glutamine synthetase (GS) gene family GS 1.705606e-02 1.127881e-01
22 NADPH oxidase (NOX) gene family NOX 1.706663e-02 1.127881e-01
26 Thaumatin-like (TLP) gene family TLP 1.954518e-02 1.237862e-01
5 Auxin transport (AUXT) gene family AUXT 2.344961e-02 1.370900e-01
28 UDP-glucuronate decarboxylase (UXS) gene family UXS 2.344961e-02 1.370900e-01
20 Phenylalanine ammonia lyase (PAL) gene family PAL 2.988103e-02 1.682191e-01
8 Core cell cycle (CCC) gene family CCC 4.239049e-02 2.301198e-01
24 S-adenosyl-L-homocysteine hydrolase (SAHH) gen... SAHH 4.544742e-02 2.382072e-01
>>> res.genfam_info
Parameter Value
0 Total query gene IDs 758
1 Number of genes annotated 460
2 Number of enriched gene families (p < 0.05) 29
3 Plant species Oryza sativa v7_JGI (Rice)
4 Statistical test for enrichment Fisher exact test
5 Multiple testing correction method Benjamini-Hochberg
6 Significance level 0.05
# check allowed ID types for rice for genfam analysis
>>> res.check_allowed_ids(species='osat')
Allowed ID types for Oryza sativa v7_JGI (Rice)
Phytozome locus: LOC_Os01g10150
Phytozome transcript: LOC_Os02g09740.2
Phytozome pacID: 24127097
In addition, you can customize different parameters for GenFam analysis (check parameters and list of plant species supported by GenFam).
GenFam citation
- Bedre R, Mandadi K. GenFam: A web application and database for gene family‐based classification and functional enrichment analysis. Plant Direct. 2019 Dec;3(12):e00191.
References
- Dametto A, Sperotto RA, Adamski JM, Blasi ÉA, Cargnelutti D, de Oliveira LF, Ricachenevsky FK, Fregonezi JN, Mariath JE, da Cruz RP, Margis R. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Science. 2015 Sep 1;238:1-2.
- Bedre R, Rajasekaran K, Mangu VR, Timm LE, Bhatnagar D, Baisakh N. Genome-wide transcriptome analysis of cotton (Gossypium hirsutum L.) identifies candidate gene signatures in response to aflatoxin producing fungus Aspergillus flavus. PLoS One. 2015 Sep 14;10(9):e0138025.
Last updated: October 10, 2020
